Noah Stier
I'm currently a senior machine learning engineer at Tesla Autopilot, working on 3D scene understanding for autonomous driving.
I did my PhD at UCSB, advised by Tobias Höllerer, where I worked on 3D reconstruction from handheld cameras. During that time I also interned with Apple's 3D reconstruction group in Seattle.
Earlier, I worked as a software engineer at Toyon and Procore, and I did my B.S. at UCLA in computational biology.

Noah Stier, Alex Rich, Pradeep Sen, Tobias Höllerer
CVPR 2025
We propose a 3D reconstruction method that adapts its resolution to the local surface complexity and orientation, achieving high surface detail very efficiently. Our surface extraction latency is reduced by nearly 25x relative to FineRecon, with almost no loss in surface accuracy.
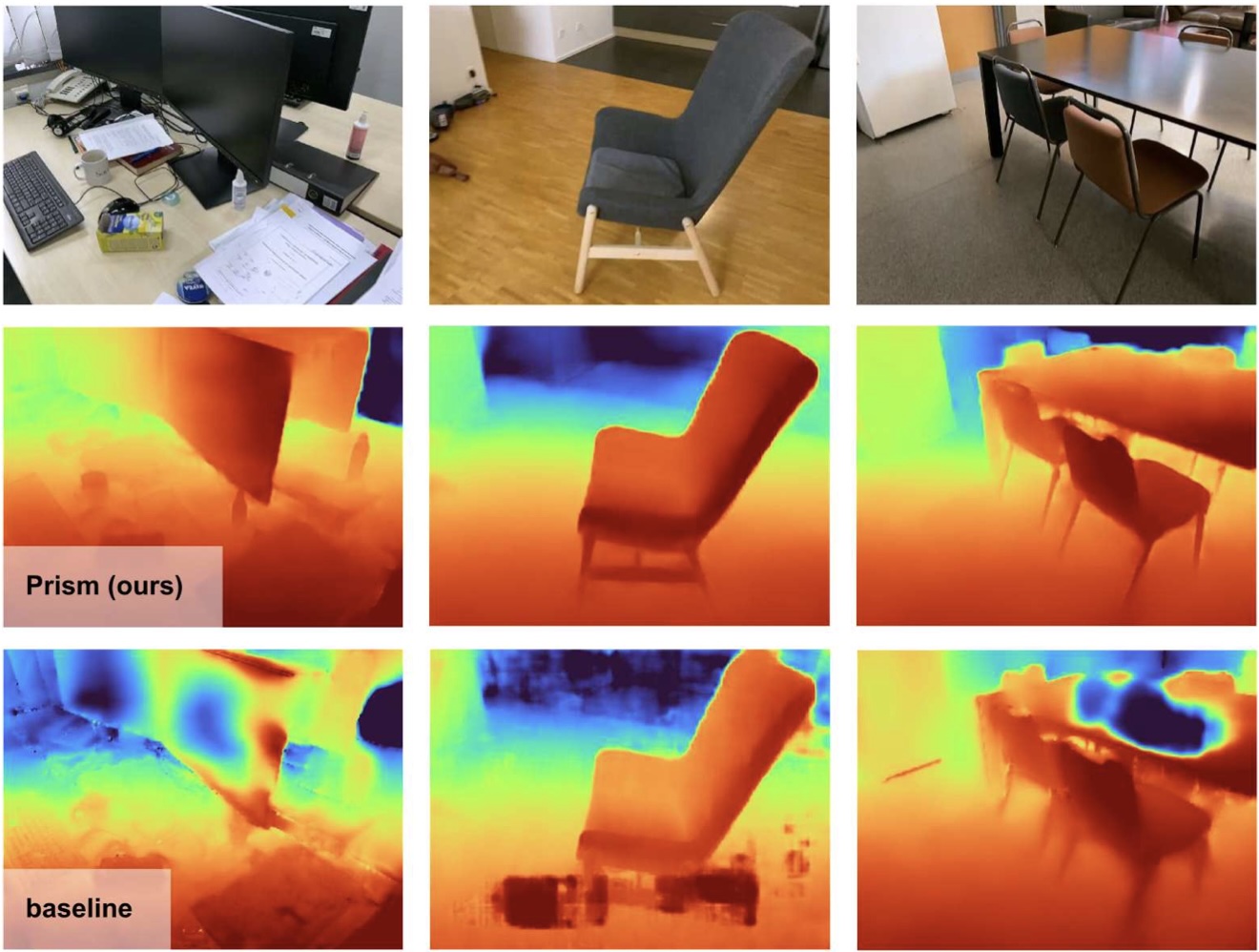
Alex Rich, Noah Stier, Pradeep Sen, Tobias Höllerer
arXiv 2025
We develop a framework for unsupervised multi-view stereo depth prediction, by distilling monocular depth priors from synthetic data, and deep feature priors from Stable Diffusion. This allows us to train highly accurate MVS networks without requiring any depth ground truth.

Chengyuan Xu, Radha Kumaran, Noah Stier, Kangyou Yu, Tobias Höllerer
ISMAR 2024
code / arXiv
We develop a vision-language 3D reconstruction system, anchoring CLIP features to the reconstructed mesh to support two augmented reality applications on the Magic Leap 2: spatial search with free-text natural language queries, and intelligent object inventory tracking.
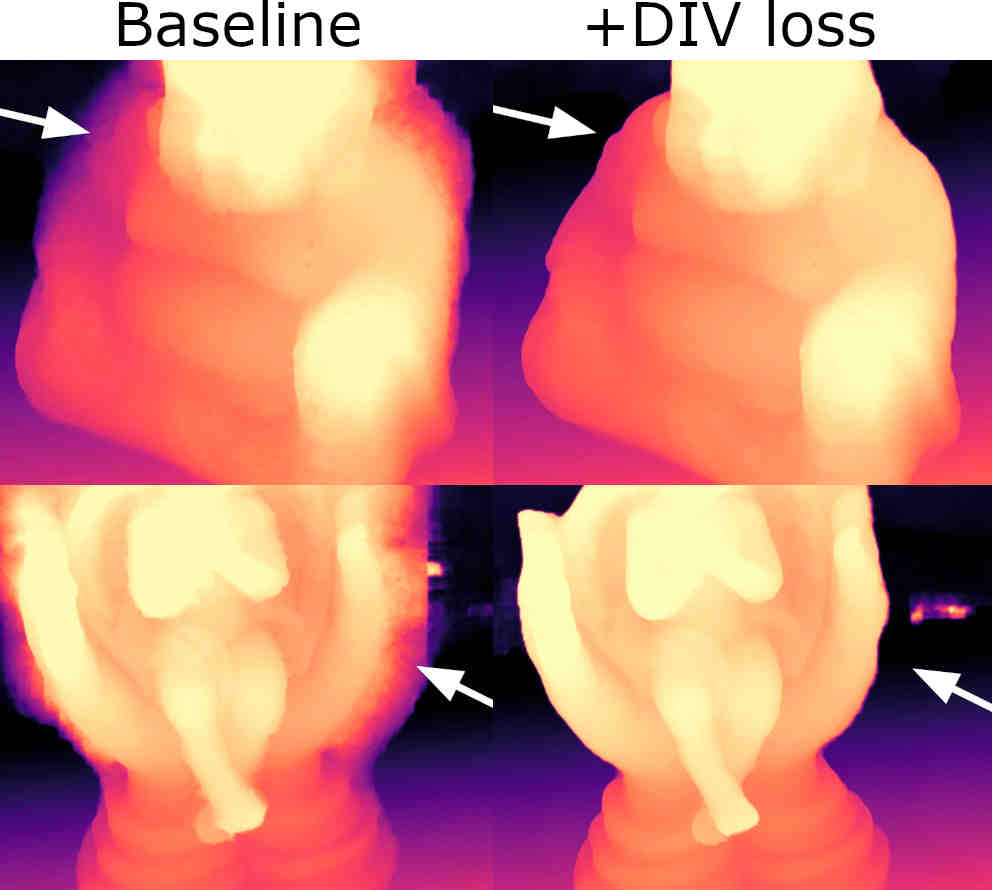
Alex Rich, Noah Stier, Pradeep Sen, Tobias Höllerer
ECCV 2024 (Oral Presentation)
We re-structure the core training objective for unsupervised MVS, allowing our networks to learn far better object boundaries for clean and coherent 3D reconstructions without requiring depth/3D ground truth.

Noah Stier, Anurag Ranjan, Alex Colburn, Yajie Yan, Liang Yang, Fangchang Ma, Baptiste Angles
ICCV 2023
code / arXiv / supplementary
We improve the reconstruction of high-frequency content by fixing a fundamental sampling error for volumetric TSDF ground truth. We also leverage depth guidance from MVS, and a new point back-projection architecture, to achieve highly accurate and detailed reconstructions.
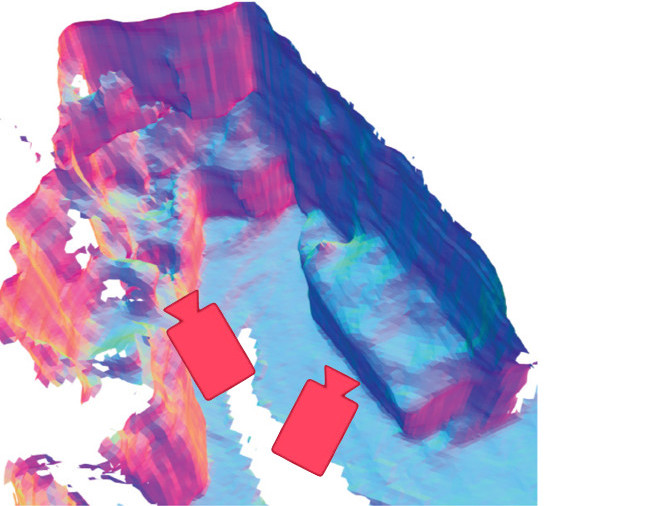
Noah Stier, Baptiste Angles, Liang Yang, Yajie Yan, Alex Colburn, Ming Chuang
ICCV 2023 (Oral Presentation)
data / arXiv
We propose neural de-integration to handle SLAM pose updates during online reconstruction from monocular video. To study this problem, we introduce the LivePose dataset of online SLAM pose estimates on ScanNet, the first to include full pose streams with online updates.
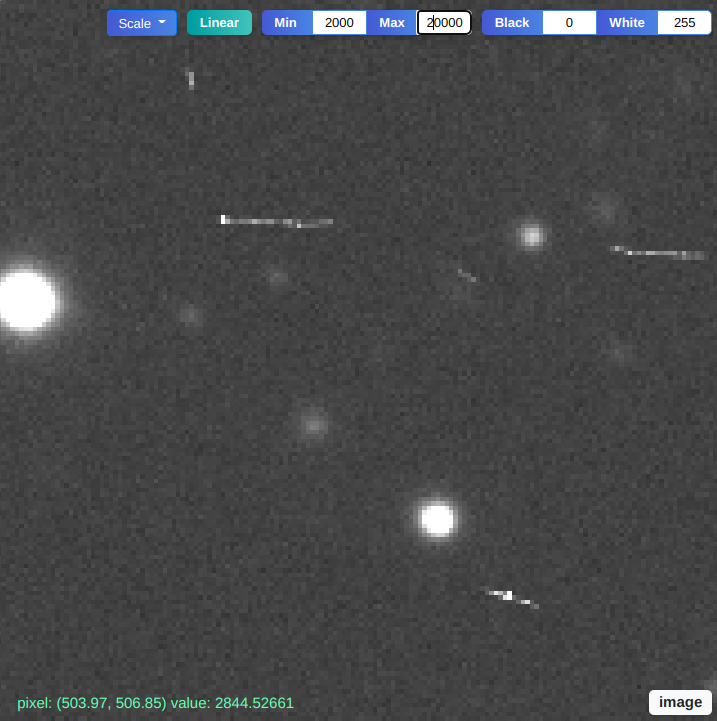
Chengyuan Xu, Boning Dong, Noah Stier, Curtis McCully, D. Andrew Howell, Pradeep Sen, Tobias Höllerer
CVPR 2022, demo track
We present an open-source software toolkit for identifying, inspecting, and editing tiny objects in multi-megapixel HDR images. These tools offer streamlined workflows for analyzing scientific images across many disciplines, such as astronomy, remote sensing, and biomedicine.
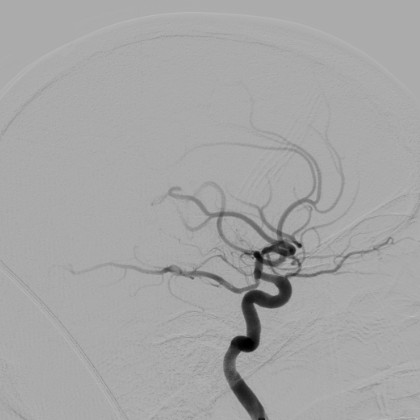
Arvind Vepa, Andrew Choi, Noor Nakhaei, Wonjun Lee, Noah Stier, Andrew Vu, Greyson Jenkins, Xiaoyan Yang, Manjot Shergill, Moira Desphy, Kevin Delao, Mia Levy, Cristopher Garduno, Lacy Nelson, Wandi Liu, Fan Hung, Fabien Scalzo
WACV 2022
We present a high-quality, annotated DSA vessel segmentation dataset, the largest to-date that is publicly available. We identify semi-automated annotation methods that significantly reduce the labeling cost while preserving the ability to train accurate segmentation models.

Noah Stier, Alex Rich, Pradeep Sen, Tobias Höllerer
3DV 2021 (Oral Presentation)
code / arXiv
We introduce a 3D reconstruction model with a novel multi-view fusion method based on transformers. It models occlusion by predicting projective occupancy, which reduces noise and leads to more detailed and complete reconstructions.

Alex Rich, Noah Stier, Pradeep Sen, Tobias Höllerer
3DV 2021
We introduce a deep multi-view stereo network that jointly models all depth maps in scene space, allowing it to learn geometric priors across entire scenes. It iteratively refines its predictions, leading to highly coherent reconstructions.
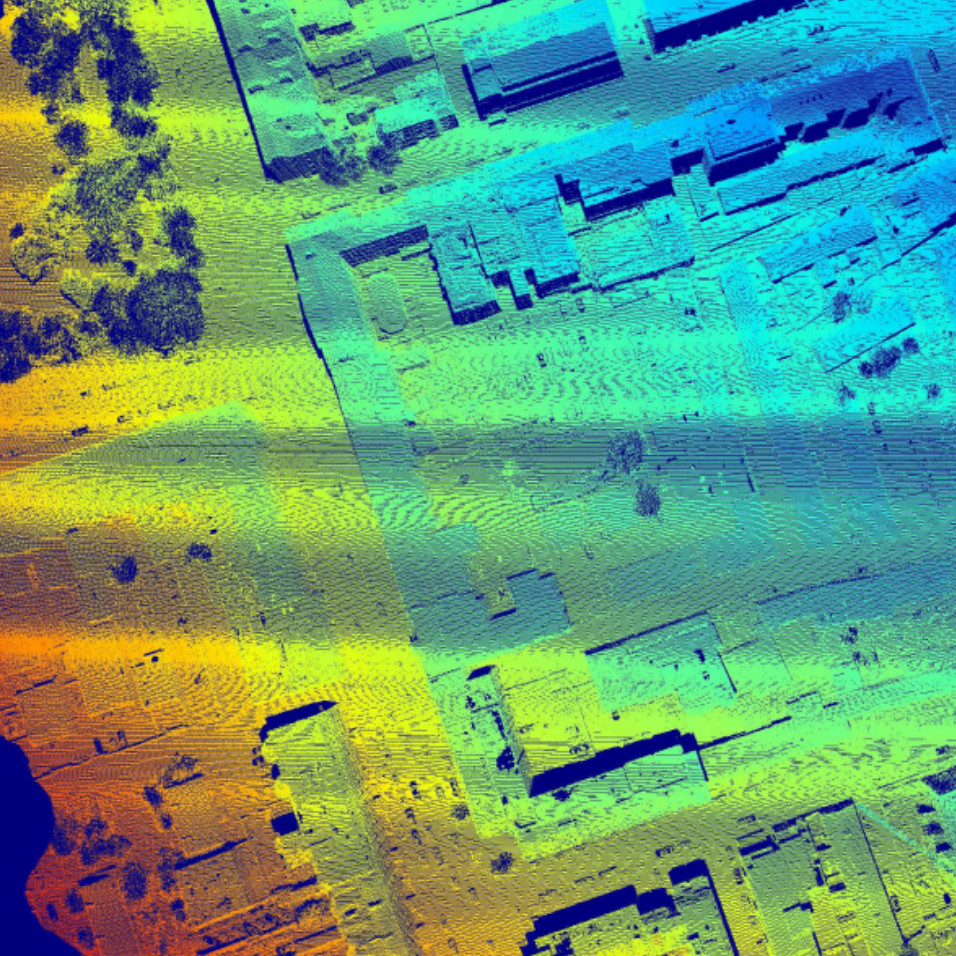
Abhejit Rajagopal, Noah Stier, William Nelson, Shivkumar Chandrasekaran,
Andrew P Brown
SPIE DCS 2020
We present a framework for wide-area detection and recognition in co-incident aerial RGB images and LiDAR, and we source ground-truth semantic labels from OpenStreetMap.

Abhejit Rajagopal, Noah Stier, Joyoni Dey, Michael A King, Shivkumar Chandrasekaran
SPIE Medical Imaging (MI) 2019
We propose deep neural networks for CT that are equivalent to classical methods at initialization, but can be trained to optimize performance while maintaining convergence properties and interpretability.
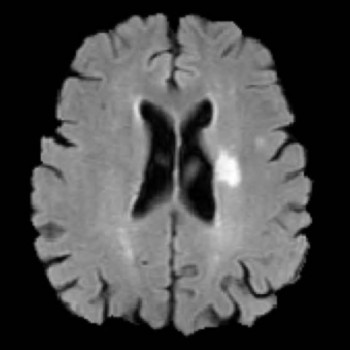
Noah Stier, Nicholas Vincent, David S Liebeskind, Fabien Scalzo
IEEE Bioinformatics and Biomedicine (BIBM) 2015
We predict the extent of tissue damage following stroke, using the cerebral hypoperfusion feature observed in MRI after stroke onset. We show improved performance using a CNN vs. the standard single-voxel regression model.
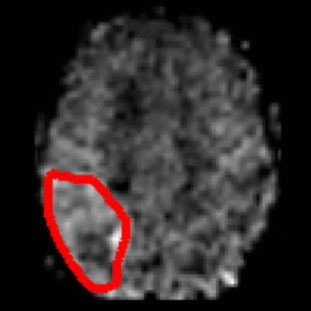
Nicholas Vincent, Noah Stier, Songlin Yu, David S Liebeskind, Danny JJ Wang, Fabien Scalzo
IEEE Bioinformatics and Biomedicine (BIBM) 2015
We train a CNN to classify regions of hyperperfusion based on cerebral blood flow (CBF) observed in MRI after stroke onset, approaching the accuracy of human experts.
Website design from Jon Barron.
